Concepts in phased assemblies
Concepts in phased assembly:
-
Contig: a contiguous sequence in an assembly. A contig does not contain long stretches of unknown sequences (aka assembly gaps).
-
Scaffold: a sequence consists of one or multiple contigs connected by assembly gaps of typically inexact sizes. A scaffold is also called a supercontig, though this terminology is rarely used nowadays.
-
Assembly: a set of contigs or scaffolds. In the following, I will say an assembly is haploid complete or simply complete if it is supposed to represent a haploid genome in full.
-
Haplotig: a contig that comes from the same haplotype. In an unphased assembly, a contig may join alleles from different parental haplotypes in a diploid or polyploid genome.
-
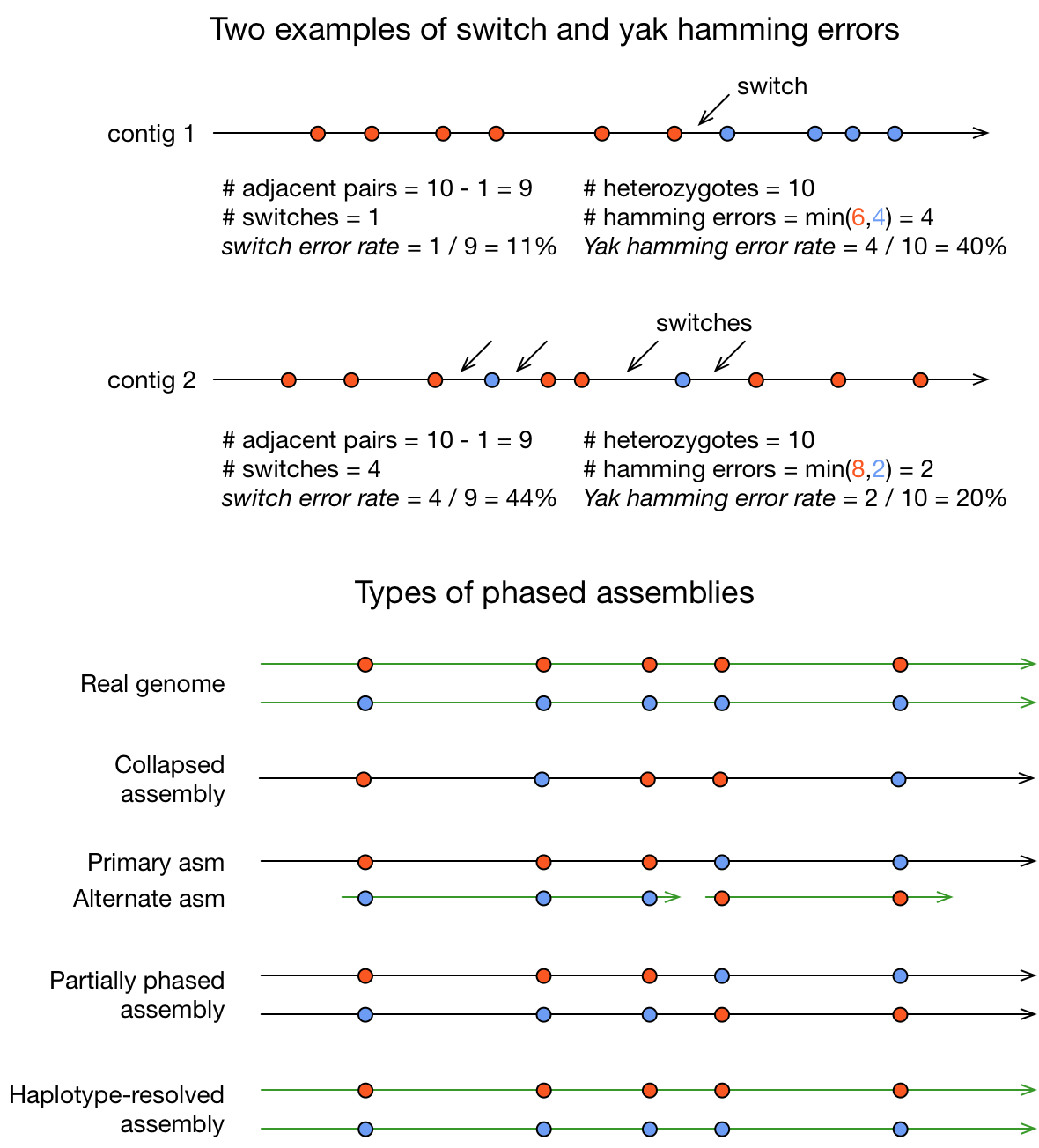
Switch error: a change from one parental allele to another parental allele on a contig (see the figure below). This terminology has been used for measuring reference-based phasing accuracy for two decades. A haplotig is supposed to have no switch errors.
-
Yak hamming error: an allele not on the most supported haplotype of a contig (see the figure below). Its main purpose is to test how close a contig is to a haplotig. This definition is tricky. The terminology was perhaps first used by Porubsky et al (2017) in the context of reference-based phasing. However, adapting it for contigs is not straightforward. The yak definition is not widely accepted. The hamming error rate is arguably less important in practice (Richard Durbin, personal communication).
Types of phased assemblies.
-
Collapsed assembly: a complete assembly with parental alleles randomly switching in a contig. Most conventional assemblers produce collapsed assemblies. A collapsed assembly is also called a squashed assembly or a conensus assembly.
-
Primary assembly: a complete assembly with long stretches of phased blocks. The concept has been used by GRC. BAC-to-BAC assemblies can all be regarded as primary assemblies. Falcon-unzip is perhaps the first to produce such assemblies for whole-genome shotgun data.
-
Alternate assembly: an incomplete assembly consisting of haplotigs in heterozygous regions. An alternate assembly always accompanies a primary assembly. It is not useful by itself as it is fragmented and incomplete.
-
Partially phased assembly: sets of complete assemblies with long stretches of phased blocks, representing an entire diploid/polyploid genome. Peregrine is perhaps the first to produce such assemblies. This concept is coined by me as I could not find a proper one in the existing literature. I don’t like the terminology. If someone has a better naming, let me know.
-
Haplotype-resolved assembly: sets of complete assemblies consisting of haplotigs, representing an entire diploid/polyploid genome. This concept has been inconsistently used in publications without a clear definition. The above is my take in our recent papers.
Furthermore, we may have chromosome-scale haplotype-resolved assembly where haplotigs from the same chromosome are fully phased. For germline genomes, the highest standard is telomere-to-telomere assembly where each chromosome is fully phased and assembled without gaps.
| Asm type | Complete? | Haplotig? | N50 | SwitchErr | HammingErr |
|---|---|---|---|---|---|
| Collapsed | Yes | No | Long | Many | Many |
| Primary | Yes | No | Long | Some | Many |
| Alternate | No | Yes | Short | Few | Few |
| Partial | Yes | No | Long | Some | Many |
| Resolved | Yes | Yes | Long | Few | Few |
blog comments powered by Disqus